LaFaC researchers promote the teaching of metabolism using metabolic models
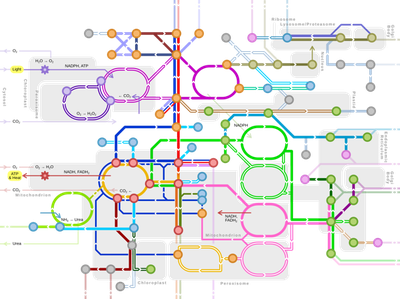
Teaching cellular metabolism to engineering students can be a challenging task, given the gap between the languages used in the biological and exact sciences. Nevertheless, a basic understanding of cellular functioning is essential for comprehending and developing fermentative processes and is part of the training of chemical and bioprocess engineers, among other professionals.
In order to bring cellular metabolism content closer to a language more familiar to Chemical Engineering students at UFSCar, LaFaC researchers used computational tools to simulate the behavior of microbial cells through mathematical models. For this purpose, the open-source software Optflux, developed by researchers at the University of Minho in Portugal, was employed to design a set of exercises aimed at studying the cellular responses of industrially relevant microorganisms under different environmental conditions.
The students involved in the project performed simulations of the metabolic models of Escherichia coli and the baker’s yeast Saccharomyces cerevisiae. Glycolysis and the Krebs cycle were among the topics covered, in addition to other metabolic pathways and the effects of different nutrients, aeration conditions, and the implementation of genetic modifications in the metabolism of the evaluated cells.
At the end, the students’ feedback on the experience was very positive, highlighting the software’s ease of use as a key factor in the success of the proposed methodology, which proved highly effective in promoting student engagement and learning.
The complete study was published in the journal Education for Chemical Engineers and can be accessed here.
.
 (2).png)